01:00
- Use keyboard arrow keys to
- advance ( → ) and
- go back ( ← )
- Type “s” to see speaker notes
- Type “?” to see other keyboard shortcuts
Part III: Transform
The Data Analysis Pipeline
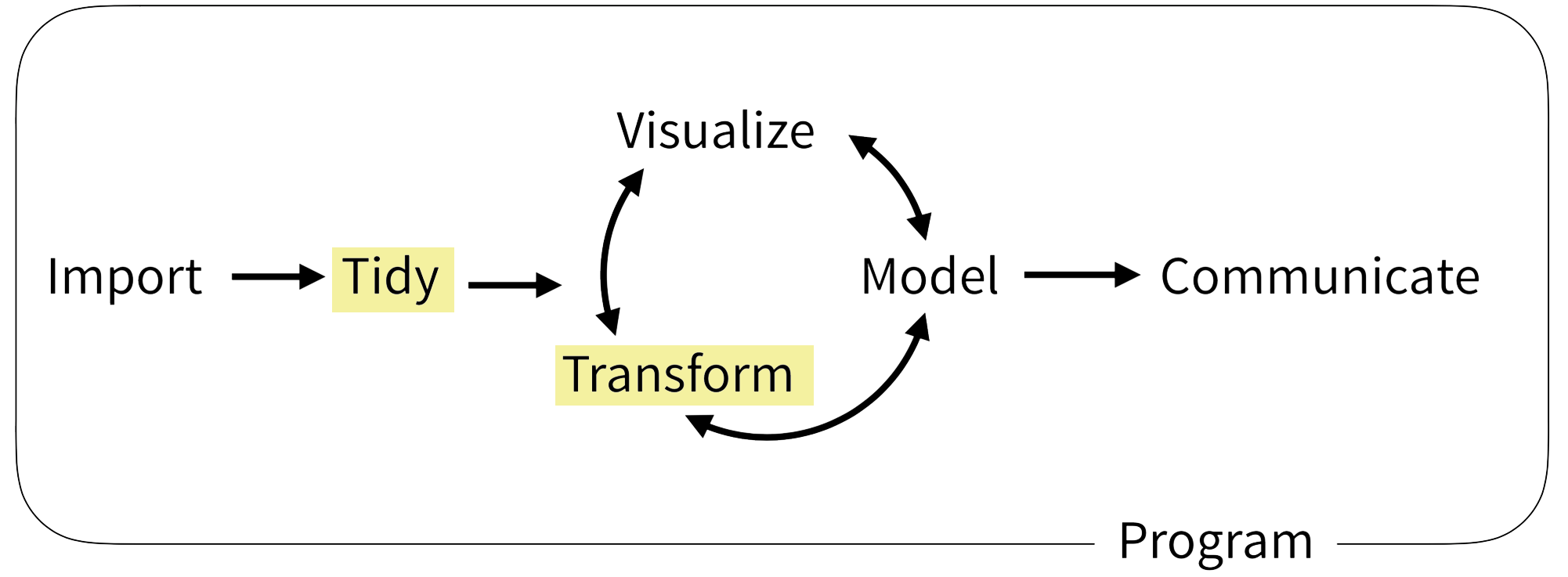

a grammar for transforming data frames
library(dplyr) OR library(tidyverse)
Subsetting Data
Subsetting Columns vs Rows
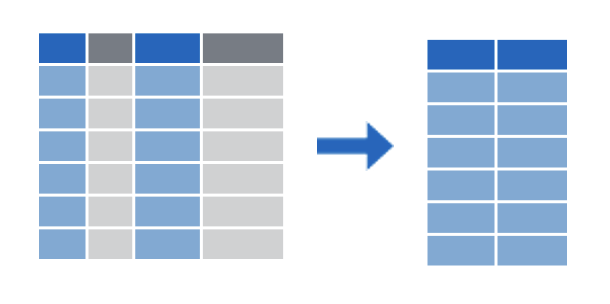
select()
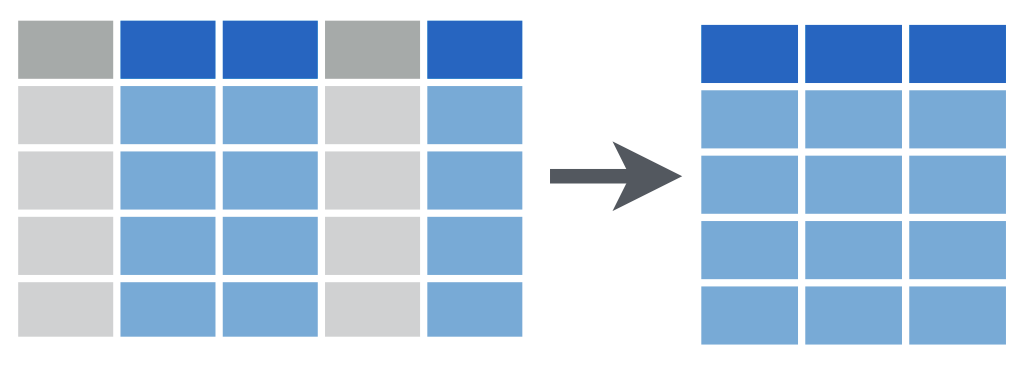
filter()
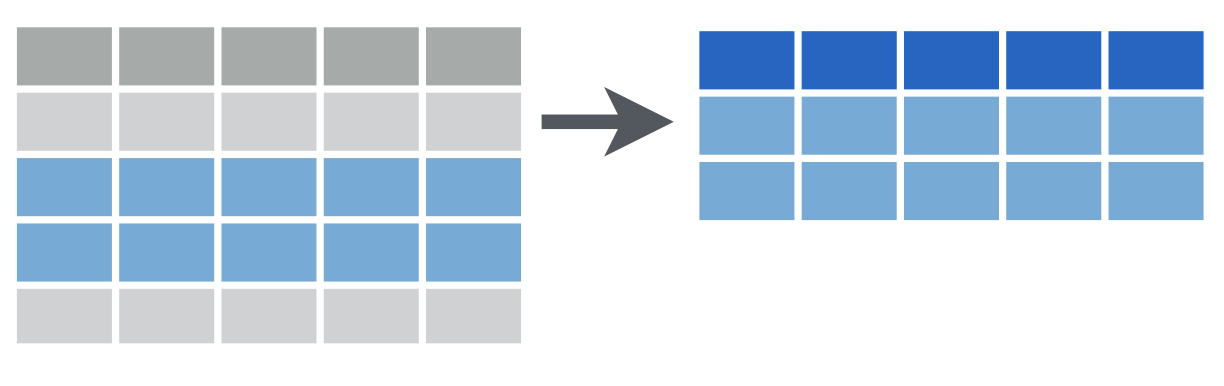
select()
select(data_frame, ...)select()
select(covid_testing, mrn, last_name)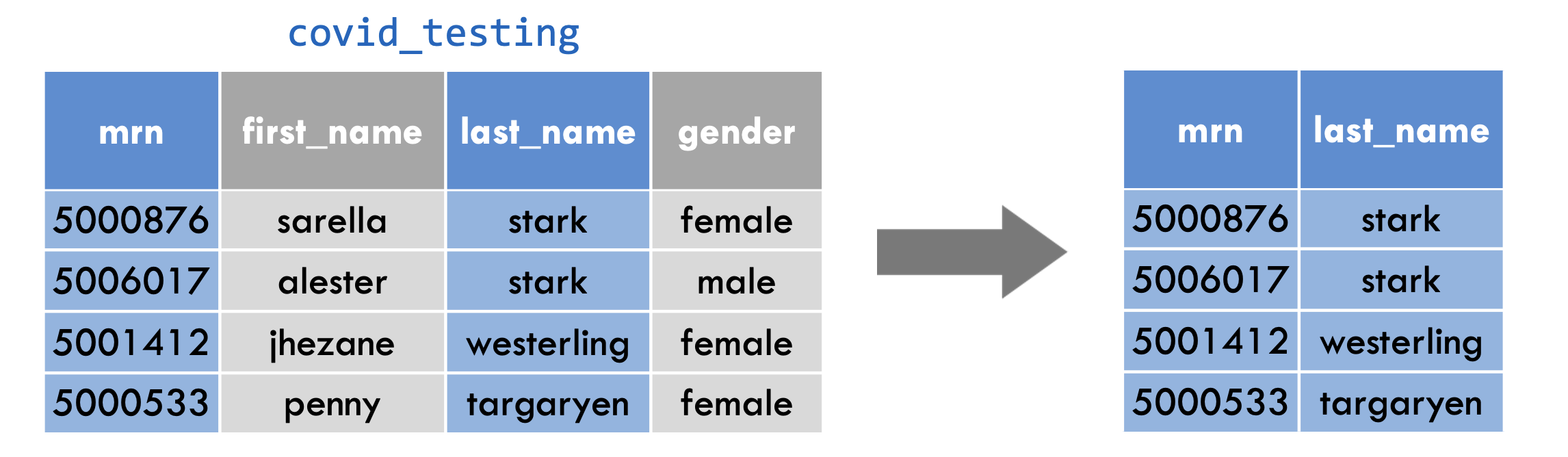
Your Turn #1
Which of the following will select the first_name column from the covid_testing data frame and capture the result in a data frame named newdata?
A) newdata = select(first_name, covid_testing)
B) newdata <- select(covid_testing, first_name)
C) select(newdata, covid_testing, first_name)
D) newdata <- select(covid_testing, First_Name)
E) Both B and D
A poll will come up for you to put in your answer in Teams!
filter()
filter(data_frame, ...)filter()
filter(covid_testing, mrn == 5000083)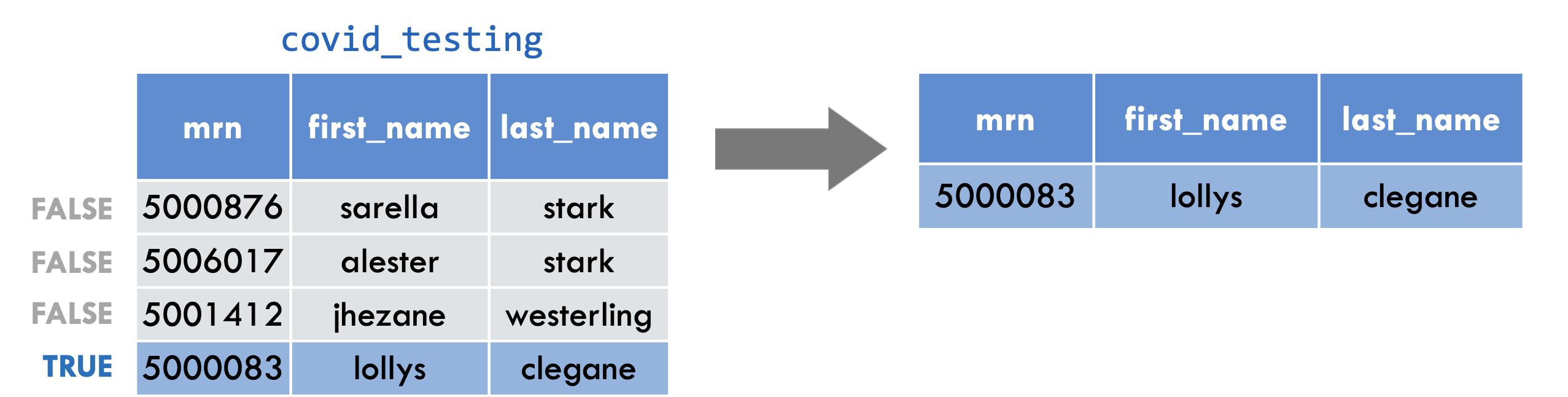
A Potential Pitfall!
Error: Problem with filter() input ..1. x Input ..1 is named. ℹ This usually means that you’ve used = instead of ==.
OR
Error: unexpected ‘=’
OR
invalid (do_set) left-hand side to assignment
Logical Operators
| logical expression | means | example |
|---|---|---|
x < y |
less than | pan_day < 10 |
x > y |
greater than | mrn > 5001000 |
x == y |
equal to | first_name == last_name |
x <= y |
less than or equal to | mrn <= 5000000 |
x >= y |
greater than or equal to | pan_day >= 30 |
x != y |
not equal to | test_id != "covid" |
is.na(x) |
a missing value | is.na(clinic_name) |
!is.na(x) |
not a missing value | !is.na(pan_day) |
Your Turn #2
Write a filter() statement that returns a data frame containing only the rows from covid_testing in which the last_name column is NOT equal to “stark”.
(You don’t have to capture the returned data frame)
Type your response in the chat!
01:00
filter(covid_testing, last_name != "stark")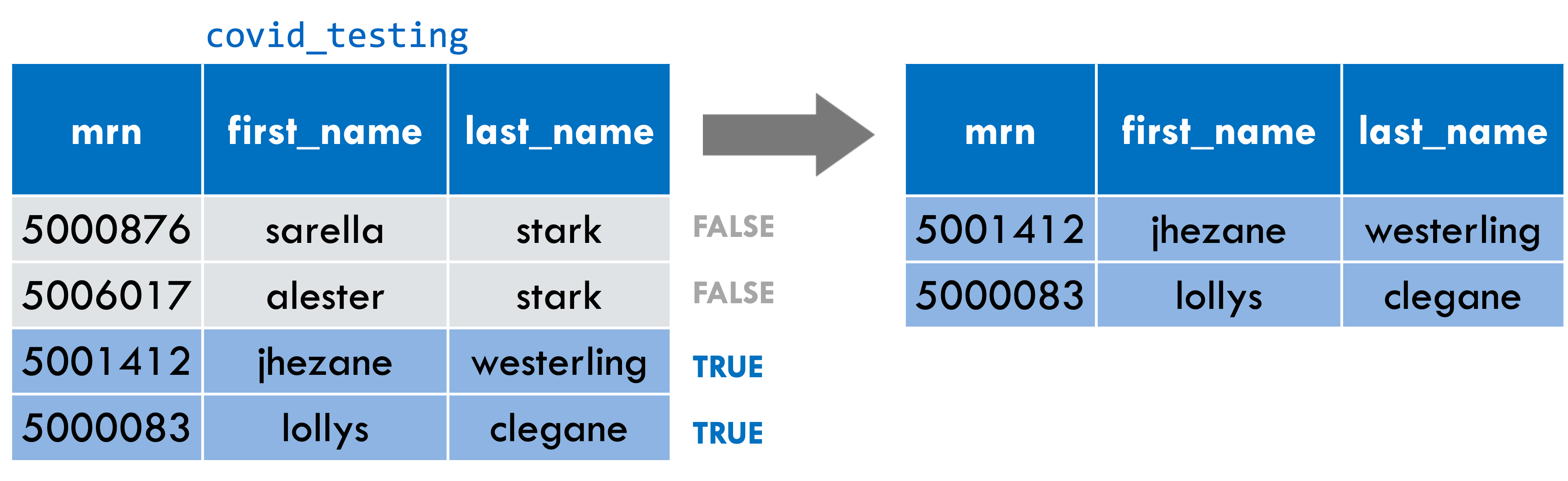
Your Turn #3
Which of these would successfully filter the covid_testing data frame to only tests with positive results?
A) filter(covid_testing, result == positive)
B) filter(covid_testing, result = “positive”)
C) filter(covid_testing, result == “positive”)
D) filter(covid_testing, positive == “result”)
01:00
The Pipe Operator %>%
The Pipe Operator %>%
The pipe operator we’ll use is %>%
(You can also use |>, in R 4.1.0 forward)
The Pipe Operator
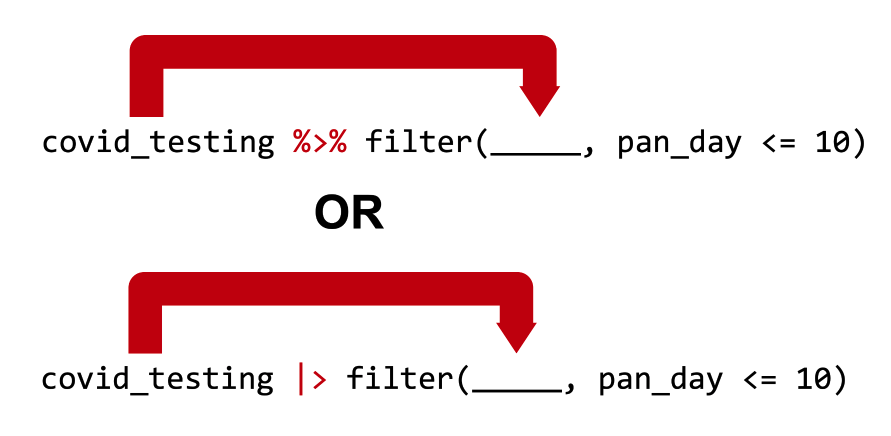
Passes the object on the left as the first argument to the function on the right
covid_testing %>% filter(pan_day <= 10) is equivalent to filter(covid_testing, pan_day <= 10)
OR, if you in the future use the “new” pipe:
covid_testing |> filter(pan_day <= 10) is equivalent to filter(covid_testing, pan_day <= 10)

- Start with the
covid_testingdata frame. THEN - Select so that we get only certain columns. THEN
- Filter so that we get only certain rows.
Your Turn #4
Rewrite the following statement with a pipe:
select(mydata, first_name, last_name)
Type the answer in the chat!
01:00
Create or Update Columns
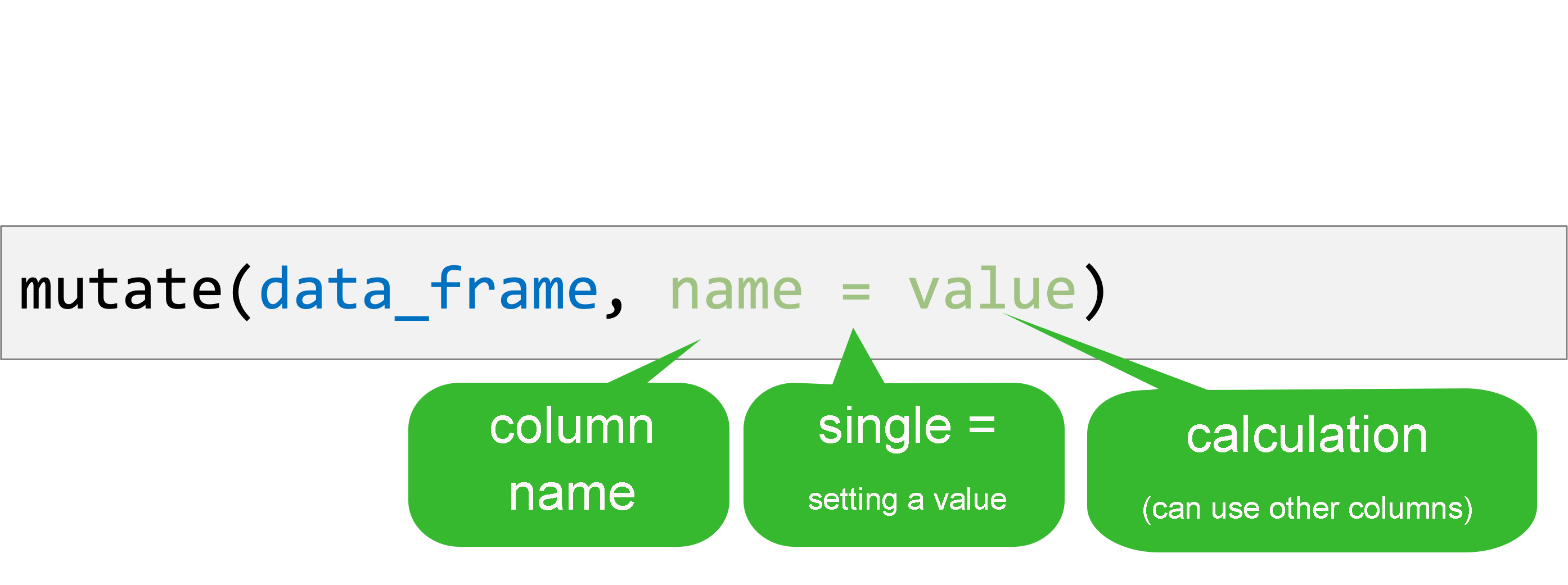
mutate()
Create new or updated, optionally calculated columns.

mutate()
Create new or updated, optionally calculated columns.

mutate()
Create new or updated, optionally calculated columns.
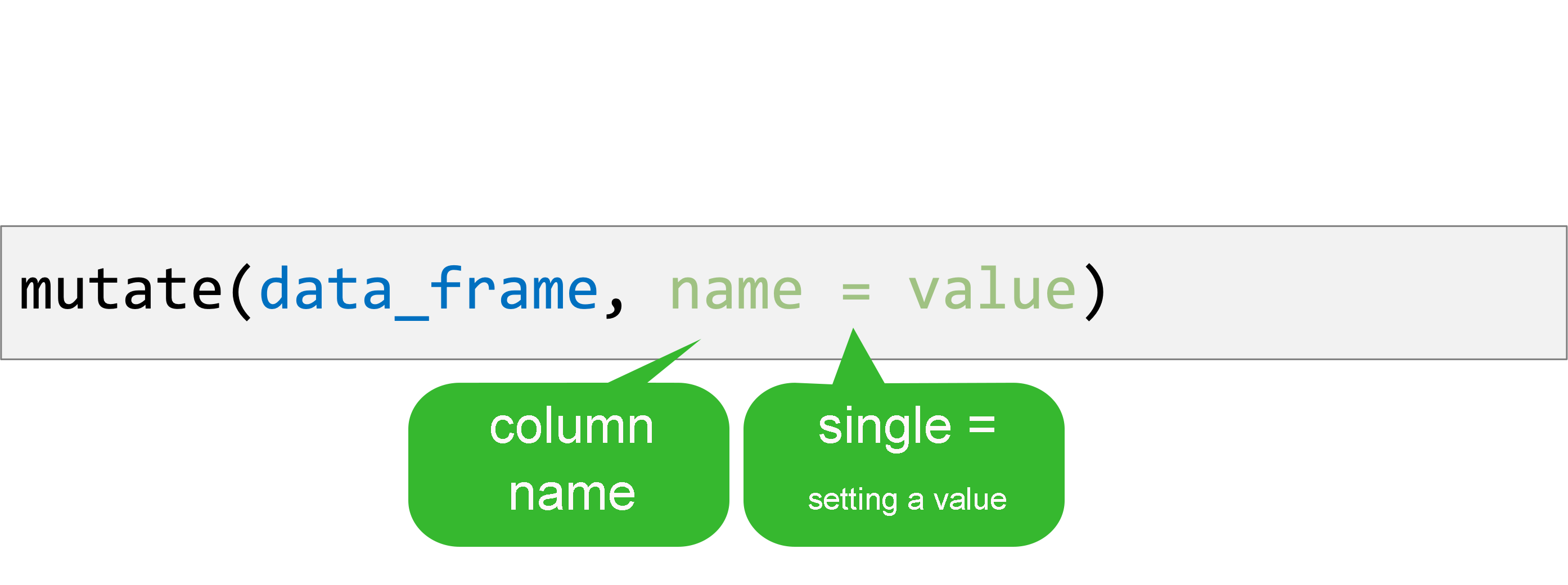
mutate()
Create new or updated, optionally calculated columns.
mutate()
mutate(covid_testing,
col_rec_tat_mins = col_rec_tat * 60)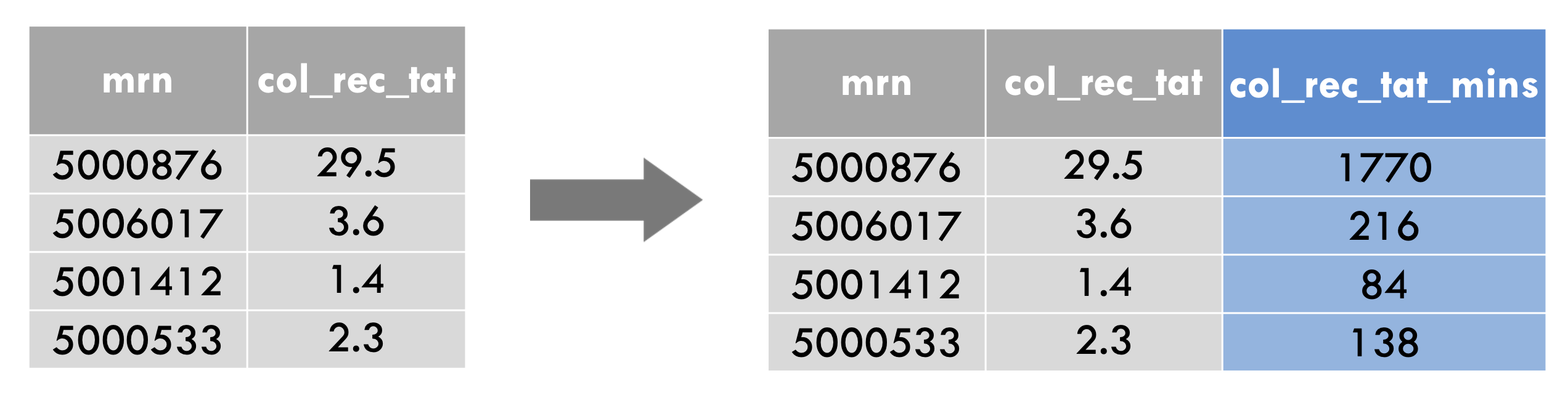
mutate()
mutate(covid_testing,
ct_value = round(ct_value))Your Turn #5
Open 03 – Transform.qmd and work through the exercises for the section that says “Your Turn #5.”
Click “thumbs up” when you are finished.
05:00
Group By and Summarize
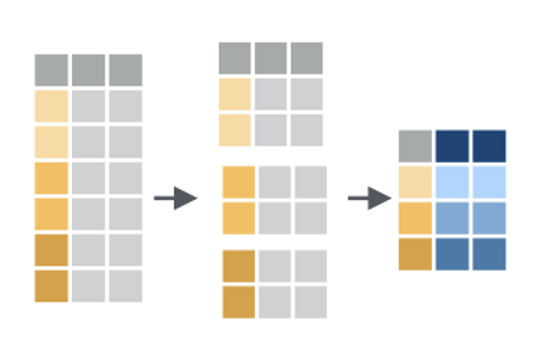
A very common use case is to divide your data into groups, and get information about each group.
For this, we’ll use group_by and summarize.
Additional Practice (Time Permitting)
If time permits:
Open 03 – Transform.qmd and work through the exercises for the section that says “Your Turn #6. We’ll do this together!
Recap
select() subsets columns by name
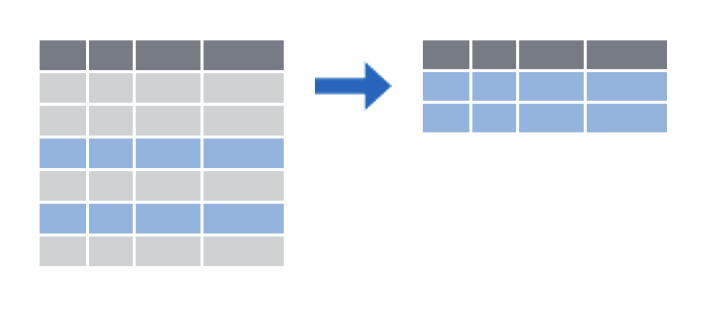
filter() subsets rows by a logical condition
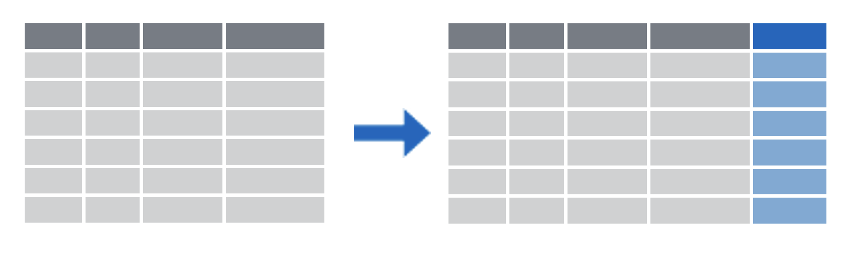
mutate() creates new calculated columns or changes existing columns
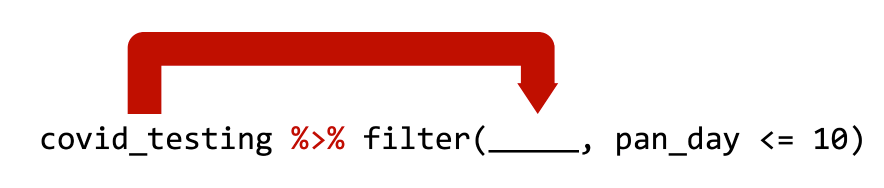
Use the pipe operator %>% to combine dplyr functions into a pipeline

group_by() with summarize() gives per-group statistics
What Else?
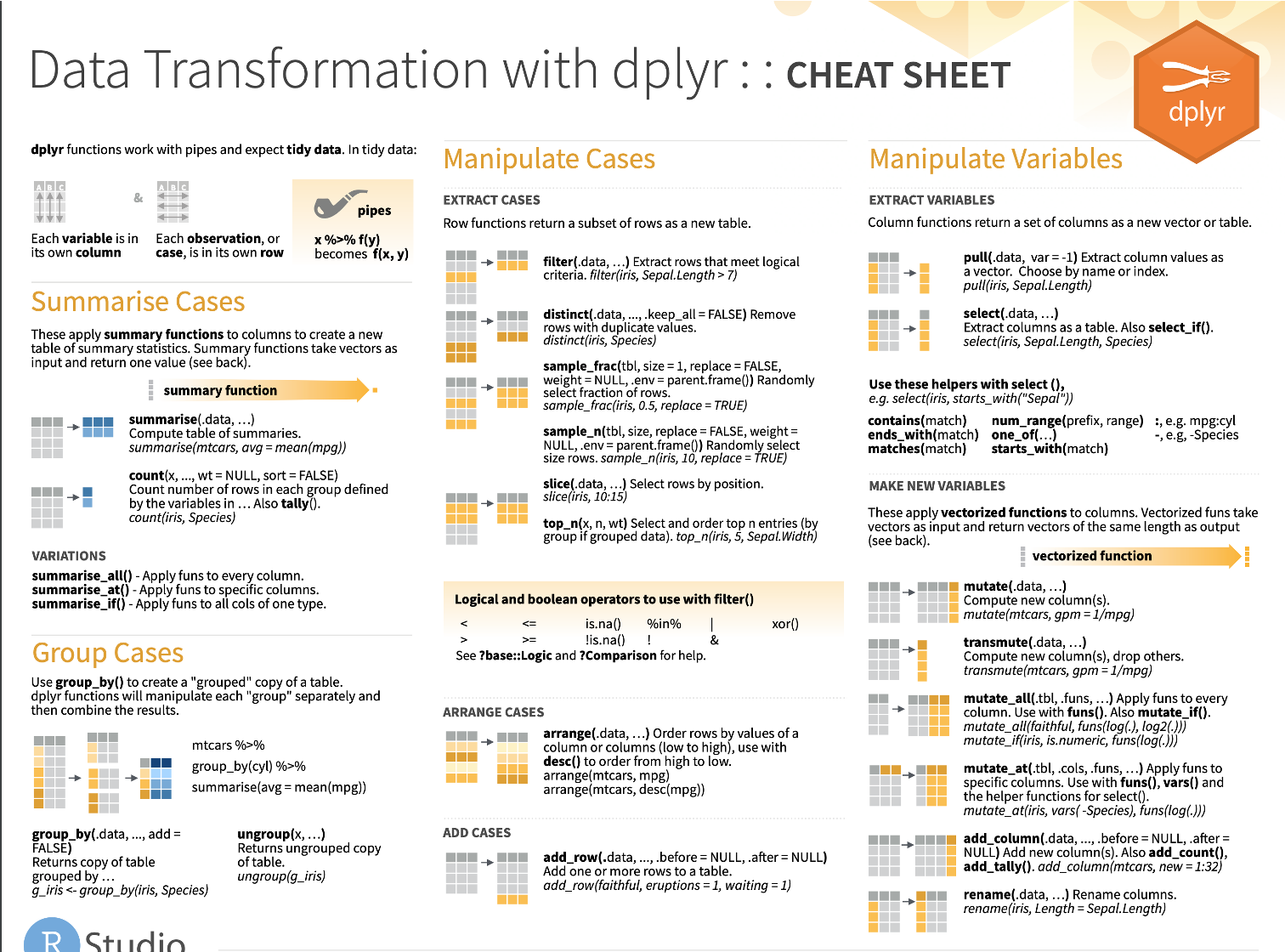
Cheatsheet (more dplyr functions!)

Next Up: Final Notes
If you want to look at Dashboards, a section we have decided to cut for time, you can find that here: Dashboards.
But we’ll be moving on to Final Notes.

Arcus Education / Children’s Hospital of Philadelphia (CHOP) R User Group